Table of contents
Preamble
The starting point of the discussion with a teammate was the performance of a SQL query in one of our application. The query in this application is running against a delta table (so in Databricks). The question was the capability of Databricks to avoid reading some of the delta files (data skipping) and ultimately to avoid reading entirely some files to only seek and read what is required (not sure on how to call it but why not row group skipping).
As delta underground is parquet columnar file I had no doubt about it but seeing it in action was the opportunity to test and understand few concepts so this blog post.
This blog post has been written using Databricks Runtime (DBR) 9.1 LTS. And even at the time of writing this post this is already old.
Data skipping test case
I have been searching a lot for the test case figures I would use as a trial. As suggested in few Databricks blogs and posts I could have used the New York City Taxi data. But I have finally decided to use something we have also sadly seen too much: the Covid 19 worldwide figures.
Upload the CSV file on your Databricks environment and load it in a dataframe with:
df1 = spark.read.format("csv").option("header", "true").option("inferSchema", "true").load("dbfs:/FileStore/shared_uploads/owid_covid_data.csv") |
You can already play with this dataframe using Spark:
import pyspark.sql.functions as F display(df1) display(df1.filter(F.col('continent')=='Europe').filter(F.col('location')=='France').filter(F.col('date').like('2022-05-30%'))) print(df1.count()) |
To see data skipping and row group in action I had to generate a delta table with multiple files which was not the default with my small dataset. Means the default maximum parquet file is bigger than my dataset.
This part was a bit tricky o setup as the session parameter is spark.databricks.delta.optimize.maxFileSize. In Databricks SQL:
%sql set spark.databricks.delta.optimize.maxFileSize=512000 set spark.databricks.delta.optimize.maxFileSize |
Or in Spark
spark.conf.set("spark.databricks.delta.optimize.maxFileSize", 512000) spark.conf.get("spark.databricks.delta.optimize.maxFileSize") |
You can also specify it at delta table creation with another parameter called delta.targetFileSize:
sql("drop table if exists default.yannick01") df1.write.format("delta").mode('overwrite').option("delta.targetFileSize",512000).saveAsTable("default.yannick01") |
What’s weird is that this parameter has absolutely no impact:
%sql
--sql("show tblproperties default.yannick01").show(truncate=False)
show tblproperties default.yannick01
+----------------------+-------+
|key |value |
+----------------------+-------+
|Type |MANAGED|
|delta.minReaderVersion|1 |
|delta.minWriterVersion|2 |
|delta.targetFileSize |512000 |
+----------------------+-------+ |
Or:
%sql
-- (sql("describe detail default.yannick01")).show()
describe detail default.yannick01
+------+--------------------+-----------------+-----------+--------------------+--------------------+-------------------+----------------+--------+-----------+--------------------+----------------+----------------+
|format| id| name|description| location| createdAt| lastModified|partitionColumns|numFiles|sizeInBytes| properties|minReaderVersion|minWriterVersion|
+------+--------------------+-----------------+-----------+--------------------+--------------------+-------------------+----------------+--------+-----------+--------------------+----------------+----------------+
| delta|93f4aeee-b1e0-4b6...|default.yannick01| null|dbfs:/user/hive/w...|2022-06-02 09:38:...|2022-06-02 09:38:30| []| 1| 12583220|{delta.targetFile...| 1| 2|
+------+--------------------+-----------------+-----------+--------------------+--------------------+-------------------+----------------+--------+-----------+--------------------+----------------+----------------+ |
Or with dbutils.fs:
dbutils.fs.ls('dbfs:/user/hive/warehouse/yannick01') Out[13]: [FileInfo(path='dbfs:/user/hive/warehouse/yannick01/_delta_log/', name='_delta_log/', size=0), FileInfo(path='dbfs:/user/hive/warehouse/yannick01/part-00000-dbaca7a1-e5ba-4d13-81fe-e99e86347942-c000.snappy.parquet', name='part-00000-dbaca7a1-e5ba-4d13-81fe-e99e86347942-c000.snappy.parquet', size=12583220)] |
My table has one datafile of size 12,583,220 bytes…
This is in fact explained in Databricks official documentation:
If you want to tune the size of files in your Delta table, set the table property delta.targetFileSize to the desired size. If this property is set, all data layout optimization operations will make a best-effort attempt to generate files of the specified size. Examples here include optimize with Compaction (bin-packing) or Z-Ordering (multi-dimensional clustering), Auto Compaction, and Optimized Writes.
So let’s ZORDER the table on continent and location with:
%sql
optimize default.yannick01 zorder by (continent, location)
path metrics
dbfs:/user/hive/warehouse/yannick01 {"numFilesAdded": 24, "numFilesRemoved": 1, "filesAdded": {"min": 374587, "max": 814246, "avg": 632810, "totalFiles": 24, "totalSize": 15187440},
"filesRemoved": {"min": 12583220, "max": 12583220, "avg": 12583220, "totalFiles": 1, "totalSize": 12583220}, "partitionsOptimized": 0,
"zOrderStats": {"strategyName": "minCubeSize(107374182400)", "inputCubeFiles": {"num": 0, "size": 0}, "inputOtherFiles": {"num": 1, "size": 12583220},
"inputNumCubes": 0, "mergedFiles": {"num": 1, "size": 12583220}, "numOutputCubes": 1, "mergedNumCubes": null}, "numBatches": 1,
"totalConsideredFiles": 1, "totalFilesSkipped": 0, "preserveInsertionOrder": false} |
So 24 new files added and 1 removed (_delta_log is the transaction log directory):
for file in dbutils.fs.ls('dbfs:/user/hive/warehouse/yannick01'): print('name=' + file.name + ',size=' + str(file.size) + ' bytes') name=_delta_log/,size=0 bytes name=part-00000-4fe40a11-4c67-47e6-bd0c-257e493bda9c-c000.snappy.parquet,size=12583220 bytes name=part-00000-7d446699-90b0-4ccc-8a0a-e23b1ca8d0fc-c000.snappy.parquet,size=514441 bytes name=part-00001-983ec08a-57da-4a97-8d1e-021ec27ca32e-c000.snappy.parquet,size=721864 bytes name=part-00002-e286b8a3-9aac-4ece-b73d-5c3f164eda22-c000.snappy.parquet,size=712834 bytes name=part-00003-b0a3564e-838e-4aa8-887f-0ce38f44adf8-c000.snappy.parquet,size=390155 bytes name=part-00004-da5d5e8e-c44a-4ab4-9e9c-66aff7f842c2-c000.snappy.parquet,size=598627 bytes name=part-00005-fcc9737b-4c27-4d2d-9499-6102c0bb47cd-c000.snappy.parquet,size=683346 bytes name=part-00006-f256fd92-380d-43f2-83a9-ce6789e5f88a-c000.snappy.parquet,size=615371 bytes name=part-00007-18e5d20a-92bb-4c0a-a263-33868df8f8d2-c000.snappy.parquet,size=743213 bytes name=part-00008-a1222ad9-f3e0-4aeb-bf05-1b7154716b50-c000.snappy.parquet,size=577178 bytes name=part-00009-6ae31a31-cbd4-437e-a0a7-d3088ebbee52-c000.snappy.parquet,size=606338 bytes name=part-00010-a5378904-6103-47fa-ab50-bb9ec4943a8c-c000.snappy.parquet,size=814246 bytes name=part-00011-b2c2953e-ec3a-4a16-913b-b88fc7069615-c000.snappy.parquet,size=693562 bytes name=part-00012-26953003-e44f-494c-a965-dd5d73b2cae6-c000.snappy.parquet,size=680122 bytes name=part-00013-6693f41b-a039-44f2-a56a-be791c0f0393-c000.snappy.parquet,size=584637 bytes name=part-00014-5f22220a-690d-4988-ac5f-faca0fc24de0-c000.snappy.parquet,size=603990 bytes name=part-00015-aa7cf66e-3579-41d5-ab75-5f15137cfdd2-c000.snappy.parquet,size=606238 bytes name=part-00016-0360de7e-8fc9-4c2c-bc53-a9248cccec3b-c000.snappy.parquet,size=592749 bytes name=part-00017-33f2de10-a8d7-47bd-b9a2-4f2e25ce0b54-c000.snappy.parquet,size=764980 bytes name=part-00018-8be3658c-e261-417b-bbf8-dd2c646b6894-c000.snappy.parquet,size=374587 bytes name=part-00019-2c248841-c75f-46c0-a3b2-e34f9733fb6b-c000.snappy.parquet,size=648834 bytes name=part-00020-50337c24-2e17-4ef6-8682-2a9578161873-c000.snappy.parquet,size=778575 bytes name=part-00021-c1b194ab-d9b0-4b34-a94d-927a50278162-c000.snappy.parquet,size=559643 bytes name=part-00022-aefc738d-ee75-465b-be74-8df86affd30c-c000.snappy.parquet,size=773971 bytes name=part-00023-6735ec1c-2ecd-4b30-b772-0b99882e71d8-c000.snappy.parquet,size=547939 bytes |
Well actually the old file is still here and to get rid off it use the VACUUM command (the set, that must not be used in production, is to accept the 0 hours retention)
%sql set spark.databricks.delta.retentionDurationCheck.enabled = false; vacuum default.yannick01 retain 0 hours; |
Data skipping and row chunk skipping testing
I have issued below query to extract Europe continent from Covid 19 information:
%sql select * from default.yannick01 where continent='Europe' |
Access to Spark UI directly from the Databricks notebook and display the associated SQL execution tab using the SQL Query number hyperlink. You see the execution plan of the query and you can access to the details of the Scan parquet files:
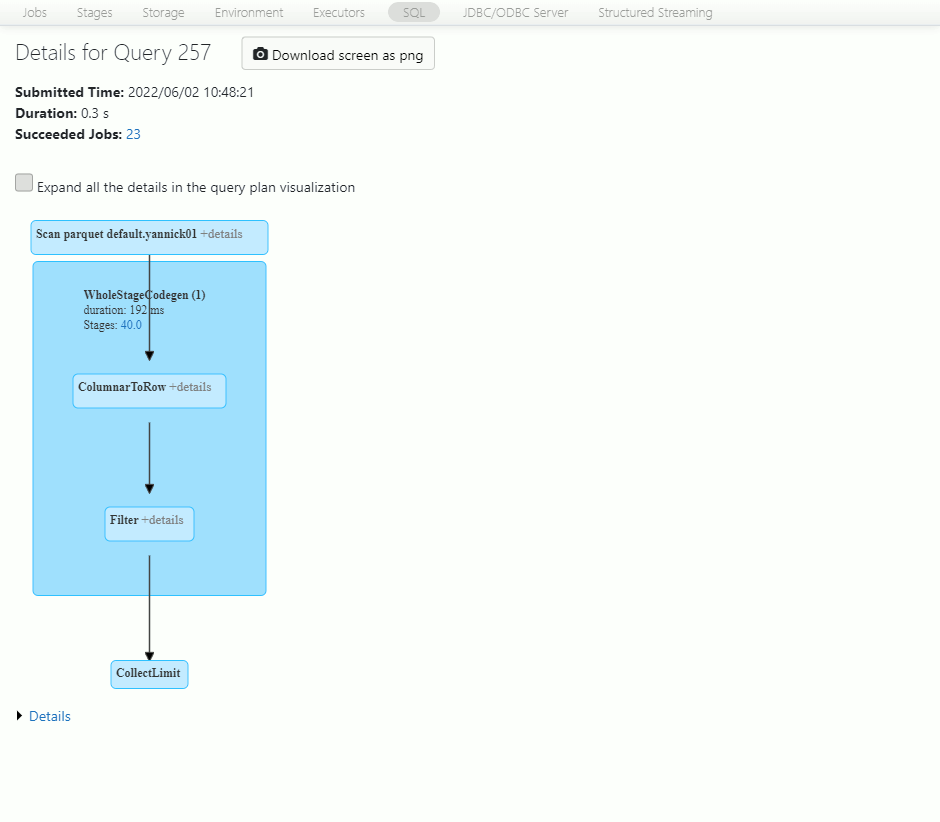
The below picture is the details of this I/O step:

What does this tell us ?
We have read all the 24 datafiles (I was not expecting this to be honest) but on the total size (size of files read) of 14.5 MB we have only read 759.2 KB (filesystem read data size) ! So we do not have data skipping but row group skipping has been fired.
I have then issued below query to extract France country from Covid 19 information:
%sql select * from default.yannick01 where location='France' |
The details of the scan parquet files step is:

This time we have read only 1 datafile of the 24 and the datafile has been entirely read, size of files read is equal to filesystem read data size. So this time data skipping has been fired while row group skipping has not been.
References
- How Parquet Files are Written – Row Groups, Pages, Required Memory and Flush Operations
- Delta table properties reference
- Processing Petabytes of Data in Seconds with Databricks Delta
- Performance Tuning Apache Spark with Z-Ordering and Data Skipping in Azure Databricks
- Delta Lake — enables effective caching mechanism and query optimization in addition to ACID properties
- Five Simple Steps for Implementing a Star Schema in Databricks With Delta Lake
- Performance enhancements in Delta Lake
- Does Spark support true column scans over parquet files in S3?
- The Parquet Format and Performance Optimization Opportunities





